We can provide homology modeling services to predict the 3D structure of proteins from amino acid sequences; On the other hand, it can provide reverse target finding service to find the possible targets of active compounds.
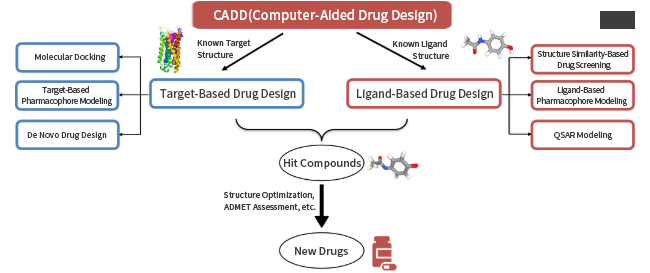
Computer Aided Drug Design (CADD) is a method based on computer chemistry to design and optimize lead compounds by computer simulation, calculation, and prediction of the relationship between drugs and recipient biomacromolecules.
Computer aided drug design is essentially to simulate the interaction between the target and the candidate drug in order to reduce the number of compounds in the solid screening experiment, so as to improve the efficiency and success rate of lead compound discovery. Random screening has an activity rate of 0.01% to 0.1%, while computer-aided drug design can quickly screen potentially active drug molecules from a large compound library (12 million topscience database stock database), and its activity rate can be increased to 5% to 20%.
Drug screens can use targeted or small-molecule structure-based drug design methods to virtually screen available compounds, natural products and other databases, and obtain a list of potentially active compounds for further activity experiments to confirm. For pharmaceutical companies and research institutes, we can provide one-stop early drug research and development services, including virtual drug screening, lead optimization, target prediction, kinetic simulation, etc., involving small molecule chemical drugs, biological drugs, traditional Chinese medicine and other new drug types, to provide you with high-quality drug discovery services.
We can provide homology modeling services to predict the 3D structure of proteins from amino acid sequences; On the other hand, it can provide reverse target finding service to find the possible targets of active compounds.
Design and optimize high quality small molecule derivatives according to experimental requirements; Small drug molecules with novel structures were found by the method of skeleton transition, and seedling compounds without intellectual rights protection were obtained

Provide molecular docking services to explore interaction patterns between protein targets and small molecules/peptides/proteins; At the same time, it provides virtual screening services for basic target/ligand structures, thereby reducing the number of screened compounds and improving the efficiency of lead compound discovery.
The ADME/T computing platform is used to predict the absorption, distribution, metabolism, excretion and toxic properties of small drug molecules in the body, reduce the elimination rate of candidate molecules, and improve the success rate of drug development.
| Disease/Medication | Target | Research Method | Active Molecule | Unit/Company |
|---|---|---|---|---|
| Senile Dementia | Beta Secretase | Virtual Filtering | For the first time, an organic small molecule inhibitor with activity of μM class was found | Shanghai Institute of Pharmaceutical Sciences |
| Arrhythmology | Potassium Channel | 3D-QSAR | A structurally novel active compound has been found, passed through animal and toxicity tests and passed on to pharmaceutical companies | Shanghai Institute of Pharmaceutical Sciences |
| Tumor | BCL-2 | Design of Virtual Screening and Combination Library | Lead compounds were found to induce apoptosis and inhibit tumor neovascularization | Institute of Medicine, Academy of Military Sciences |
| Nervous System | FKBP12 | Design of Virtual Screening and Combination Library | The activity of 10 compounds was higher than that of the positive control FK-506 | Institute of Medicine, Academy of Military Sciences |
| Arthritis | Phospholipase A2 | De Novo Design, Virtual Screening, 3D-QSAR | Find compounds with novel structures and activities comparable to positive compounds | Department of Chemistry, Peking University |
| Acquired Immune Deficiency Syndrome | HIV Proteolytic Enzymes | The drug Saquinavir for the treatment of the human immunodeficiency virus | Roche (Welwyn, UK) | |
| Influenza | Neuraminidase | The selective antiviral drug zanamivir | Biota (Melbourne, Australia) |
Drug Screen provides customers with 12 million solid compounds, ensuring that customers can buy their ideal solid compounds after virtual screening. Will not be screened for good structure, but no inventory and worry.
Most of the compounds can provide 5mg, 10mg, 20mg, 50mg and other packaging, but also according to customer needs to provide DMSO solution packaging, most of the product delivery only 2-3 weeks, quickly meet the customer's scientific research requirements.
Drug Screen has the most complete range of molecular docking software available for virtual screening (DOCK, AutoDock, GOLD, GLIDE, Surflex, LigandFit, etc.).
Drug Screen also has other world-class commercial drug molecular design software, including MOE, Discovery Studio, SYBYL, Schrodinger and Gaussian 09, as well as a number of academic software (including AMBER, GROMACS, CHARMm, etc.).
These software can be used to complete the molecular simulation computing tasks involved in drug molecular design, such as protein homologous structure modeling, pharmacophore induction, molecular dynamics simulation, quantum chemical calculation, etc.
At present, the company has two sets of Baode high performance computing cluster, a total of 25 compute nodes (440 logical cpus), more than 15 TB of hard disk storage space. These hardware resources can be used to provide external virtual filtering services.
In order to meet the needs of biochemistry customers for computer-aided virtual screening, drug screen can help customers complete virtual screening based on molecular docking and pharmacophore based virtual screening. All the products we screen are actually available small molecule compounds, to avoid customers due to improper database selection, resulting in the target molecules can not be obtained, reduce your input of human, material and financial costs.
For most projects, we can provide a full range of services from virtual screening to the purchase of small molecule compounds to the verification of the activity of compounds to the verification of cytology experiments.